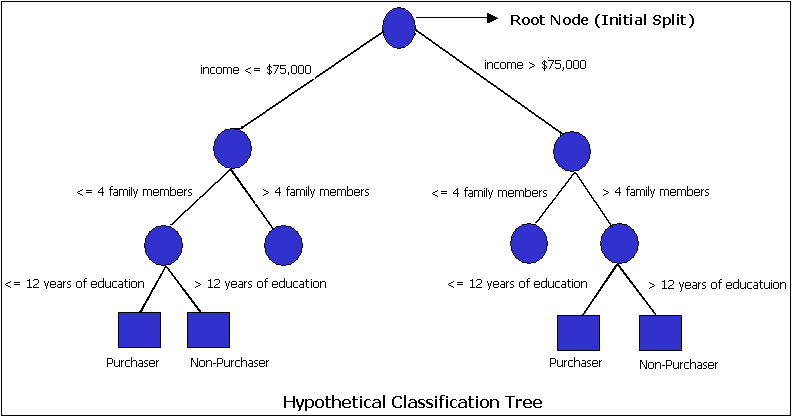
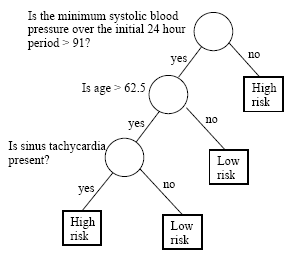
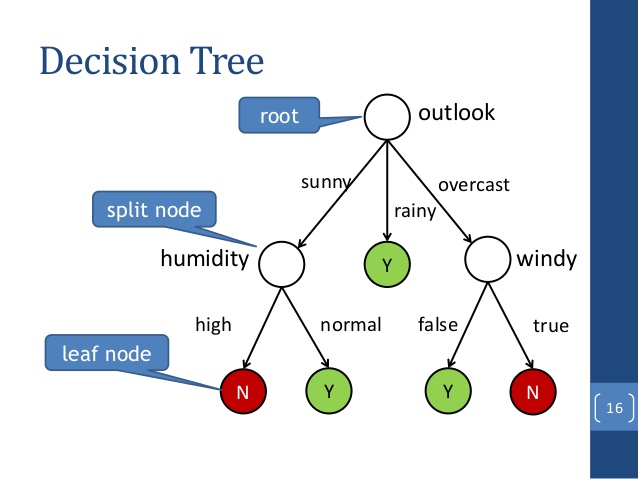
Understanding data set
We use the inbuilt data set iris.
It has 150 observations and 5 variables. We need to build a model to predict the categorical target variable 'Species' using the numerical feature variables Petal.Length, Petal.Width, Sepal.Length, Sepal.Width
# check dimensions of data set
dim(iris)
# [1] 150 5
names(iris)
#[1] "Sepal.Length" "Sepal.Width" "Petal.Length" "Petal.Width" "Species"
str(iris)
#'data.frame': 150 obs. of 5 variables:
# $ Sepal.Length: num 5.1 4.9 4.7 4.6 5 5.4 4.6 5 4.4 4.9 ...
# $ Sepal.Width : num 3.5 3 3.2 3.1 3.6 3.9 3.4 3.4 2.9 3.1 ...
# $ Petal.Length: num 1.4 1.4 1.3 1.5 1.4 1.7 1.4 1.5 1.4 1.5 ...
# $ Petal.Width : num 0.2 0.2 0.2 0.2 0.2 0.4 0.3 0.2 0.2 0.1 ...
# $ Species : Factor w/ 3 levels "setosa","versicolor",..: 1 1 1 1 1 1 1 1 1 1 ...
Load installed R packages
libs = c("party")
lapply(libs, require, character.only=TRUE)
Create train and test data set
Split the data set in to two parts - 70% for training and remaining 30% for testing
Use sample function to shuffle the dataset and assign index 1 to 70% of data and index 2 to remaining data points; such that index 1 can be used as training set and 2 as testing set
#sample the data by assigning index positions
ind = sample(2, nrow(iris), replace=TRUE, prob=c(0.7,0.3))
str(ind)
# int [1:150] 2 1 1 1 1 1 2 1 2 1 ...
# train dataset is all rows with index 1, 70% of dataset
trainData = iris[ind==1,]
str(trainData)
# 'data.frame': 96 obs. of 5 variables:
# $ Sepal.Length: num 4.9 4.7 4.6 5 5.4 5 4.9 5.4 4.8 5.8 ...
# $ Sepal.Width : num 3 3.2 3.1 3.6 3.9 3.4 3.1 3.7 3.4 4 ...
# $ Petal.Length: num 1.4 1.3 1.5 1.4 1.7 1.5 1.5 1.5 1.6 1.2 ...
# $ Petal.Width : num 0.2 0.2 0.2 0.2 0.4 0.2 0.1 0.2 0.2 0.2 ...
# $ Species : Factor w/ 3 levels "setosa","versicolor",..: 1 1 1 1 1 1 1 1 1 1 ...
# test dataset is all rows with index 2, 30% of dataset
testData = iris[ind==2,]
str(testData)
# 'data.frame': 54 obs. of 5 variables:
# $ Sepal.Length: num 5.1 4.6 4.4 4.8 4.3 5.1 5.1 4.6 4.8 5 ...
# $ Sepal.Width : num 3.5 3.4 2.9 3 3 3.8 3.7 3.6 3.4 3 ...
# $ Petal.Length: num 1.4 1.4 1.4 1.4 1.1 1.5 1.5 1 1.9 1.6 ...
# $ Petal.Width : num 0.2 0.3 0.2 0.1 0.1 0.3 0.4 0.2 0.2 0.2 ...
# $ Species : Factor w/ 3 levels "setosa","versicolor",..: 1 1 1 1 1 1 1 1 1 1 #...
Cross check the prediction model
Since the model is built using training data, it should predict target values for training set accurately
# checking prediction model with the train data
trainPred = predict(iris_ctree, data=trainData)
# confusion matrix for predicted and actual values
table(trainPred, trainData$Species)
#trainPred setosa versicolor virginica
#setosa 35 0 0
#versicolor 0 28 0
#virginica 0 4 29
Plot the classification tree
plot(iris_ctree)
Use the model to predict test data target
Here we predict the Species for the test data using the classification tree model that was built using the training data
# now use the ctree model to predict Species for test dataset
testPred = predict(iris_ctree, newdata=testData)
testPred
# [1] setosa setosa setosa setosa setosa
# [6] setosa setosa setosa setosa setosa
# [11] setosa setosa setosa setosa setosa
# [16] versicolor versicolor versicolor versicolor versicolor
# [21] versicolor virginica versicolor versicolor virginica
# [26] versicolor versicolor versicolor versicolor versicolor
# [31] versicolor versicolor versicolor virginica virginica
# [36] versicolor virginica virginica virginica virginica
# [41] virginica virginica virginica virginica virginica
# [46] virginica virginica virginica virginica virginica
# [51] virginica virginica virginica virginica
# Levels: setosa versicolor virginica
str(testPred)
# Factor w/ 3 levels "setosa","versicolor",..: 1 1 1 1 1 1 1 1 1 1 ...
Determine accuracy of model
Based on the predictions on test data accuracy is calculated as the mean of correct predictions for the target variable Species
#calculate accuracy of prediction
accuracy = mean(testPred == testData$Species)
accuracy
#[1] 0.9444444
error = mean(testPred != testData$Species)
error
#[1] 0.05555556

